AI Research
Canadian scientists pioneer made-in-Canada quantum-powered AI solution to solve worldwide challenge

Newswise — WATERLOO, ON and VANCOUVER, BC – In a landmark achievement for Canadian science, a team of scientists led by TRIUMF and the Perimeter Institute for Theoretical Physics have unveiled transformative research that – for the first time – merges quantum computing techniques with advanced AI to model complex simulations in a fast, accurate and energy-efficient way.
“This is a uniquely Canadian success story,” said Wojciech Fedorko, Deputy Department Head, Scientific Computing at TRIUMF. “Uniting the expertise from our country’s research institutions and industry leaders has not only advanced our ability to carry out fundamental research, but also demonstrated Canada’s ability to lead the world in quantum and AI innovation.”
Traditional simulations of particle collisions are already both time-consuming and costly, often running on massive supercomputers for weeks or months. By leveraging quantum processes and technology made possible by California-based D-Wave Quantum Inc., the researchers were able to create a new “quantum-assisted” generative model capable of running simulations and open new opportunities to cost-effectively analyze rapidly growing data sets.
The research, published today in npj Quantum Information, is part of a worldwide effort to create the tools needed to accommodate upgrades to CERN’s particle accelerator, the Large Hadron Collider (LHC), and alleviate a computational bottleneck that would impact researchers all over the world.
“Our method shows that quantum and AI technologies developed here in Canada can solve real-world scientific bottlenecks,” said Javier Toledo-Marín, joint appointee at TRIUMF and Perimeter Institute. “By combining deep learning with quantum technology, we are forging a new path for both theoretical experimentation and technological application.”
In addition to TRIUMF and Perimeter, contributions to the published research came from the National Research Council of Canada (NRC), the University of British Columbia and the University of Virginia, showcasing not only the wealth of research talent and scientific ingenuity across the country, but also the international collaboration that places Canada at the forefront of worldwide scientific innovation.
-30-
For more information:
For Perimeter:
Mark Healy
For TRIUMF:
Stuart Shepherd
About Perimeter Institute for Theoretical Physics
Perimeter Institute is the world’s largest research hub devoted to theoretical physics, fostering breakthroughs in the fundamental understanding of our universe, from the smallest particles to the entire cosmos. For 25 years, Perimeter’s innovative approach has empowered scientists worldwide to explore emerging scientific fields, giving researchers the freedom they need to question assumptions and blaze new trails. Research at Perimeter is motivated by the understanding that fundamental science advances human knowledge and catalyzes innovation, and that today’s theoretical physics is tomorrow’s technology. Founded by visionary Mike Lazaridis in 1999 and located in the Region of Waterloo, the not-for-profit Institute is a unique public-private endeavour, including the Governments of Ontario and Canada, that enables cutting-edge research, trains the next generation of scientific pioneers, and shares the power of physics through award-winning educational outreach and public engagement.
About TRIUMF
Established in 1968 in Vancouver, TRIUMF is Canada’s particle accelerator centre. The lab is a hub for discovery and innovation inspired by a half-century of ingenuity in answering nature’s most challenging questions. From the hunt for the smallest particles in our universe to research that advances the next generation of batteries or develops isotopes to diagnose and treat disease, TRIUMF drives more than scientific discovery. Powered by its complement of top talent and advanced accelerator infrastructure, TRIUMF is pushing the frontiers in isotope science and innovation, as well as technologies to address fundamental and applied problems in particle and nuclear physics, and the materials and life sciences.
AI Research
College Of Engineering Introduces Artificial Intelligence Degree

The School of Electrical Engineering and Computer Science (EECS) in Penn State’s College of Engineering announced an artificial intelligence engineering (AIE) degree option opening this fall.
As one of the first artificial intelligence-centered programs in the country, the course content will focus on foundations and applications of AI, hardware, and software, as stated in a release from the college.
In the release, Tom La Porta, the director of the School of EECS, explained that the degree differs from other AI programs due to the combination of computer science, computer engineering, and electrical engineering.
“This degree covers a much wider range and deeper set of topics than most AI degrees, ranging from hardware, through algorithms to applications and implementation,” he stated.
Along with the undergraduate degree option, a minor in AIE is also available for students who wish to incorporate it into their chosen field of study. According to the college, the minor provides scholars with AI techniques that can be particularly useful for computer science, computer engineering, and electrical engineering majors.
Penn State Hartz Family Associate Professor of Computer Science and Engineering Mehrdad Mahdavi was one of the faculty members who helped create the degree from scratch, according to the release.
“Our AIE degree is not just learning a bunch of tools and applying it. Students will apply their knowledge and experience to address applications of AI to complex problems. Additionally, they will gain a deeper understanding of the issues required to develop responsible AI that benefits individuals, organizations, and society,” Mahdavi said.
Mahdavi explained that the AIE program is structured in three levels: it begins with AI foundations, then builds on that knowledge through the second level, and culminates at the top layer filled with electives covering different disciplines.
To learn more about the AIE major or minor, contact Mahdavi at [email protected] or Jack Sampson at [email protected].
AI Research
Oxford, Ellison Institute Launch AI Vaccine Research

Led by the Oxford Vaccine Group , the new initiative based in the University – CoI-AI (Correlates of Immunity-Artificial Intelligence) – will combine Oxford’s expertise in human challenge studies, immune science and vaccine development with EITs cutting edge Artificial Intelligence (AI) innovation technology to better understand how the body fights infection and how vaccines protect us.
The CoI-AI programme will study how the immune system responds to important germs that cause serious infections and contribute to antibiotic resistance – such as Streptococcus pneumoniae, Staphylococcus aureus, and E. coli, – amongst others, which cause widespread illness but have resisted traditional vaccine approaches. Researchers will use human challenge models (where volunteers are safely exposed to bacteria under controlled conditions) and apply modern immunology and AI tools to pinpoint the immune responses that predict protection.
Professor Sir Andrew Pollard , Director of the Oxford Vaccine Group , said: ‘This programme addresses one of the most urgent problems in infectious disease by helping us to understand immunity more deeply to develop innovative vaccines against deadly diseases that have so far evaded our attempts at prevention. By combining advanced immunology with artificial intelligence, and using human challenge models to study diseases, CoI-AI will provide the tools we need to tackle serious infections and reduce the growing threat of antibiotic resistance. This is a new frontier in vaccine science.’
Professor Daniela Ferreira , Deputy Director of the Oxford Vaccine Group, said: ‘This programme will give us completely new tools to study how vaccines work at both a cellular and system-wide level, by studying infections in real time, in people, and using smart immunology tools and data to find the answers. This will open up whole new avenues to vaccine design as we improve our understanding of infection and immunity.’
 Professor Daniela Ferreira
Professor Daniela FerreiraLarry Ellison, Chairman of the Ellison Institute of Technology , said: ‘Researchers in the CoI-AI programme will use Artificial Intelligence models developed at EIT to identify and better understand the immune responses that predict protection. This vaccine development programme combines Oxford’s leadership in immunology and human challenge models with cutting-edge AI, laying the groundwork for a new era of vaccine discovery – one that is faster, smarter, and better able to respond to infectious disease outbreaks throughout the world.’
Professor Irene Tracey, Vice-Chancellor of the University of Oxford, said: ‘This is a major step forward in our strategic alliance with the Ellison Institute. Together, we’re combining Oxford’s strengths in vaccine science with EIT’s bold vision to tackle some of the toughest problems in global health. This is about drawing more talent and capacity to the Oxford ecosystem to turn scientific challenges into real solutions for the world.’
In December 2024 the University of Oxford and EIT announced a long-term strategic alliance to develop the solutions and future leaders needed to tackle some of the greatest and most enduring challenges facing humanity.
EIT combines cutting-edge research and commercial capability, to drive scientific breakthroughs and create sustainable, ethical companies, bringing together cross-cutting capabilities and talent spanning generative biology, clinical medicine, plant science, sustainable energy, public policy, and more.
These efforts are underpinned by substantial computing capability enabled by Oracle, a world-class Artificial Intelligence team, and a Scholars programme to develop future generations of the world’s finest scientists.
The Oxford Vaccine Group is a vaccine research group within the Department of Paediatrics at the University of Oxford’s Medical Sciences Division .
AI Research
Mitigating antimicrobial resistance by innovative solutions in AI (MARISA): a modified James Lind Alliance analysis
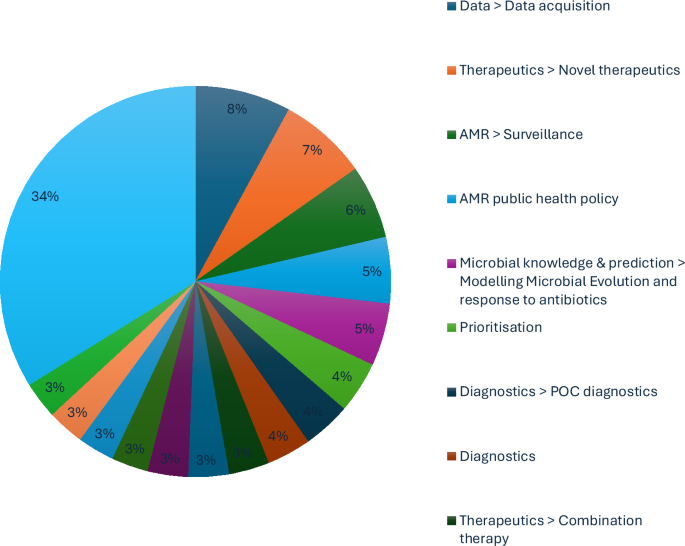
These results have set out the direction of expert opinion for AMR policy, particularly in how to optimise research and development efforts. Nevertheless, there remain obstacles to explore in implementing this expert vision.
Brokered data-sharing was proposed by the expert interviews to unify the divided efforts against AMR. A data broker is a regulator and diplomat; a referee of how data is collected and used, but also an unbiased arbitrator between commercially competitive organisations. This precedent is established in other industrial contexts by the UK Information Commissioner’s Office11 and European Data Protection Regulation12. Data was the most consistently raised issue throughout the interviews. The interviewees were aware that any machine-learning algorithm often requires large amounts of data, with concerns centred around access to datasets that already exist, particularly pharmacological data rather than microbial data. Possible solutions include a federated pharmacological dataset that does not compromise participating commercial research programmes, such as through a data broker. This would require standardisation of data collection or at least a data cleaning methodology. Moreover, Wan et al. demonstrated that AI models can be trained with less data than previously thought; APEX was trained on standardised experimental data collected for about 1000 compounds tested against an array of target bacteria, yielding a matrix of about 15,000 data points13.
AI-driven modelling is essential for the development of novel therapeutic options. Creating novel therapeutics was seen as a very high priority by interviewees, though most mentioned the importance of pursuing multiple avenues, as there is significant uncertainty as to where the future lies. Safety must be a central theme and concern. Alongside specifically targeting drugs, modelling pharmacokinetics and pharmacodynamics could also be approached using AI methods. Peptides, immunologics and small molecules were all seen as reasonable targets14,15,16. The paucity of anti-fungal therapy was highlighted as a pressing issue. Phage was viewed favourably in terms of a second-line therapy or personalised medicine, but concerns were raised about their longevity as a therapeutic option and the potential for unintended consequences (e.g. acting as a vector for AMR spread).
Rapid diagnostics emerged as a key pillar in the fight against AMR. Interviewees emphasised the need for more accessible, affordable and rapid diagnostic tools that could be implemented at various levels of healthcare, including in low-resource settings. Advances in AI and molecular diagnostics can help enable real-time pathogen detection, reducing delays in treatment and limiting unnecessary antimicrobial use. Improved diagnostics would enhance antimicrobial stewardship, enable more precise epidemiological tracking and improve patient outcomes by ensuring the right treatment is administered early. Acquisition of this data needs to be standardised to optimise international efforts, which was also a subject of intense scrutiny during the AMR Data Symposium at The London School of Hygiene and Tropical Medicine in October 202417. Data is currently stored in silos corresponding to traditional disciplines, and therefore, the datasets are incomplete, whilst there remain integration issues due to heterogeneity of data collection methods from different labs or disciplines. This is corroborated by a review of antimicrobial learning systems (ALSs), which noted the obstacles of incomplete data capture, dwindling pipelines of new antimicrobials, one-size-fits-all antimicrobial treatment formularies, resource pressures and poorly implemented diagnostic innovations18. Moreover, gaps in the currently available data require further research (e.g. pharmacological studies in humans), and data is being missed due to poor testing capability, resulting in skewed datasets. The specific priority for applying genomic data in the fight against AMR was also highlighted by the Wellcome conference on AMR in March 202419. Comprehensive datasets are needed to fully realise the opportunities AI presents in combating AMR20.
Drug discovery is essential to deliver regional biosecurity. Vaccines were highly rated by interviewees, but they acknowledged the difficulty of creating anti-bacterial rather than anti-viral vaccines. Nevertheless, it is appealing to target organisms that drive high antimicrobial use. Therapeutic approaches were an area most interviewees felt was a priority, though it tended to be highlighted by those who were currently working in this field, and they admitted their bias due to their specialisation. The concept of automating preclinical drug discovery was seen as very appealing. High-throughput screening was detailed as a way that AI could be used to speed up screening of vaccine candidates and to highlight new potential mechanisms for drug targets. Combination therapies were described as a way that AI was well-suited to the large-scale screening required for finding combination therapies. However, the challenges raised by interviewees, which mostly centred around pharmacokinetic and pharmacodynamic concerns, should be noted. However, although a drug may work synergistically with another in theory, the concentrations required for antimicrobial drugs, rather than other non-antimicrobials, are often greater by an order of magnitude, which presents safety concerns. AI was postulated to model the factors that affect drug penetration, the effect of the immune system and the effect of the gut microbiome.
Integrated Economic Prevention refers to the strategic investment in preventative public health measures that mitigate financial and societal costs associated with AMR before they escalate into more severe economic and healthcare burdens. It underscores the importance of cost-effective interventions, such as improved stewardship, surveillance and public awareness campaigns, that not only reduce long-term expenditures for healthcare systems but also enhance societal engagement by demonstrating tangible financial savings. Economic prevention is an essential component of framing the public narrative. The cost of AMR and the savings to be made for the public purse through pre-emptive action are needed for improving societal engagement. Despite the ongoing research and predicted professional concern, AMR does not have the public profile of other diseases, such as cancer.
This BARDI framework for directing AMR policy is corroborated by leading reviews in this space. To that end, Rabaan et al.21 discuss how AI can address AMR by optimising antibiotic prescriptions, predicting resistance patterns and enhancing diagnostics; they emphasise the potential of machine learning to improve clinical decisions and reduce the misuse of antibiotics. Moreover, Howard et al.18 propose a framework for implementing AI-driven ALSs, and highlight AI’s role in optimising antimicrobial use, addressing the complexity of AMR data and promoting adaptive, sustainable interventions within healthcare systems; the development of ALSs is especially important for modelling in surveillance efforts and understanding the drivers of AMR22. Meanwhile, Ali et al.23 outline AI’s potential in antimicrobial stewardship, emphasising its application in predicting bacterial resistance, optimising treatment pathways and identifying emerging threats through large-scale data analysis. Furthermore, Rabaan et al.21, in alignment with the BARDI framework, focus on AI’s application in clinical settings, particularly in improving antibiotic stewardship. Moreover, Ali et al.23 further support these views by detailing AI’s role in data-driven antimicrobial stewardship, reinforcing the importance of large-scale data integration and machine learning for proactive AMR management. Moreover, inappropriate antibiotic prescribing leads to increased AMR, patient morbidity and mortality. Clinical decision support systems that integrate AI with widely used clinical screaming systems can aid clinicians by providing accurate, evidence-based recommendations. This would enable more objective prescription of antibiotics in compliance with guidelines as per diagnosis. To this end, Howard et al.18 expand on this by proposing a systemic framework for integrating AI within healthcare, stressing the importance of continuous learning systems. The obstacles to progress in AI use for AMR include incomplete or biased data, especially from primary care, which hinders accurate AI predictions. Sparse data and fragmented health systems complicate AI’s effectiveness18,20; the BARDI framework provides a basis for international cooperation against AMR.
Moreover, this BARDI framework aligns with Chindelevitch et al.24 who describe AMR advancements in surveillance, prevention, diagnosis and treatment; this research group has developed the first WHO-endorsed catalogue of mutations in the Mycobacterium tuberculosis complex associated with drug resistance and also developed INGOT-DR, an interpretable machine-learning approach to predicting drug resistance in bacterial genomic datasets25. The molecular mechanisms of AMR have been reviewed to deliver an overview of intervention and modelling opportunities, particularly using genomic sequencing and functional genomic approaches, including CRISPR26. Machine-learning applications to whole-genome studies27 present opportunities for both AMR surveillance as well as downstream drug discovery. Whilst applying AI to small molecules may generate recommendations which are not chemically synthesizable28, future models may be able to incorporate pharmacology and toxicity to deliver promising molecules such as Guavanin-2, which expressed the novel and non-coded mechanism of hyperpolarization of the bacterial membrane29. Meanwhile, with a growing demand for microbial resistance data collection, the irregular standards and practices of reporting globally compromise confidence in the field’s advancement30. Modelling AMR transmission may enable personalised antimicrobial stewardship interventions, particularly in carbapenem-resistant Klebsiella pneumoniae31. This necessitates a whole-system approach32 with collaborations such as the Surveillance Partnership to Improve Data for Action on Antimicrobial Resistance [SPIDAAR], funded by Pfizer and the Wellcome Trust33, for national plans for upscaling surveillance and prevention programmes in collaborations with private sector resources, particularly for deriving mechanistic insights on drug activity from population-scale experimental data34.
This BARDI framework is an urgent strategy to fight AMR; in a study of 41.6 million US hospital admissions (over 20% of national hospitalisations annually) between 2012 to 2017, the incidence of extended-spectrum beta-lactamase increased by 53.3% (from 37.55 to 57.12 cases per 10,000 hospitalisations), demonstrating that most hospitalisations were now showing characteristics of AMR10. If this trajectory does not stop, even the wealthiest nations will be overwhelmed.
There are at least four ongoing considerations to the implementation of this BARDI framework. Data privacy and security are important when dealing with sensitive patient information. Global standardisation and interoperability of AI systems are necessary to ensure that AI-driven AMR detection can be effectively integrated for biosecurity across different healthcare environments35. Now that 460 academic, commercial and public data consumers in the UK National Health Service have been mapped, and the challenges of multistage data flow chains have been shown to include noncompliance with best practice, there remain concerns about delivering the full potential of UK health data36, including the establishment of a data broker.
Subtle differences in the definitions of a particular clinical event can have a dramatic impact on prediction performance. Cohen et al.37, when applying tree-based, deep learning and survival analysis to the MIMI-III intensive care admissions database, demonstrated a 0-6% variation in area under the receiver operator characteristic (AUROC) depending on the onset of time in different definitions of sepsis. This is an essential observation to consider when using AI in the forecast of AMR because of the potential for resistance to be expressed in different patterns across different bacteria, as each resistant strain manifests harm to the host. A more precise definition beyond ‘when microbes no longer respond to antimicrobial medicines’38 is needed to minimise the variation in AUROC of AMR predictive diagnostics; there must not be a repeat of the error of the Sepsis-III definition in not defining the clinical event onset time39,40. Moreover, it is important to differentiate suspicion of infection and organ dysfunction when interpreting the clinical scenario41; verifiable methods with a precise definition of AMR are needed to direct a valid clinical response. Li et al.42 may have begun to address this problem by cataloguing some of the exact alleles associated with specific resistance patterns, but this would need to be improved to real-time analysis to allow for meaningful improvements to clinical decision-making.
Both ‘One Health’ and ‘Global Health’ are interdisciplinary concepts based on the interdependence of human and animal health, and integrating biological, environmental and socioeconomic factors, but they address AMR at different levels. Hospitals, patients and infections are all sometimes described as ‘resistant’43, but technically only the bacteria become resistant to an antibiotic38. However, when interrogating genomic and EHR data to develop a forecast, one is inclined to define AMR by a broader criterion of observed phenomena as the infection takes effect in the host patient and hospital. Gene capture strategies44 and real-time PCR procedures45 may improve the detection of antibiotic-resistant bacteria, regardless of their environment. This matters for a ‘Global Health’ perspective since novel resistance arises in one place and then disseminates worldwide46.
Modern vaccine platforms such as mRNA, protein subunit and vector-based technologies offer new opportunities for developing targeted vaccines against resistant pathogens, alleviating the AMR pressure on healthcare systems worldwide47. Vaccines offer a complementary approach to antimicrobials by decreasing antibiotic demand; vaccines against Streptococcus pneumoniae and Haemophilus influenzae type b have reduced rates of pneumonia and meningitis48. This reduces demand for broad-spectrum antibiotics, slowing resistance from antibiotic regimens selecting for resistant strains49. Moreover, vaccination against viral pathogens can help reduce the incidence of secondary bacterial infections and reduce symptomatic presentations, which may be confused with bacterial pathology50. In populations with high influenza vaccination coverage, there is a consequent reduction in antibiotic prescriptions, particularly during flu seasons51. mRNA vaccines, vector-based vaccines and protein subunit vaccines have expanded the potential for targeting AMR directly52. mRNA vaccines use messenger RNA to induce cells to produce specific antigens, thereby triggering an immune response to AMR pathogens53. This technology allows for the rapid development and testing of vaccines against AMR bacteria with rapid turnover, such as Pseudomonas aeruginosa and Mycobacterium tuberculosis54. Protein subunit vaccines enable targeted responses against resistant bacterial strains, such as MRSA55. Conjugate vaccines, which combine a protein with a polysaccharide to enhance immunogenicity, can prevent infections that commonly exhibit antibiotic resistance; the pneumococcal conjugate vaccine has not only reduced pneumococcal disease prevalence but also curbed the spread of drug-resistant S. pneumoniae strains56. However, caution is required as some drug-resistant strains have evaded vaccination57 and general resistance has been reported to bounce back to pre-vaccine levels58. Vector-based vaccines can be engineered to target specific resistant pathogens, especially those with complex life cycles59. Escherichia coli, Klebsiella pneumoniae and Acinetobacter baumannii are emerging targets for pharmaceutical research60 since these infections are difficult to treat due to multidrug resistance.
Moreover, these developments complement combination machine-learning methodologies, which leverage data-driven models in synthetic gene circuit engineering61; the introduction of RhoFold+62, an RNA language model-based deep learning method that accurately predicts 3D structures of single-chain RNAs from sequences, enables the fully automated end-to-end pipeline for RNA 3D structure prediction. Vaccines could significantly impact AMR by preventing infections that are notoriously hard to manage with available antibiotics. Whilst we wait for vaccines to be developed, resistant microorganisms will need to be scrutinised for druggable vulnerabilities, such as through collateral sensitivity induction by manipulating bacterial physiology63. These intermediate efforts can be supported by antimicrobial resistant gene databases64,65,66,67,68,69, particularly which offer spatiotemporal and abundancy data70.
The full James Lind Alliance (JLA) protocol process was modified; each Priority-Setting Partnership (PSP) would normally consist of patients, carers and their representatives, and clinicians, and is led by a Steering Group. The Steering Group oversees the activities of the PSP and has responsibility for the activity and the outcomes of the PSP. Since the role of the PSP is to identify questions that have not been answered by research to date, this study followed a modified path with the experts giving answers within a thematic framework. While the study excluded direct public involvement in its priority setting, it acknowledges that public awareness and engagement are critical in combating AMR. Instead of broad public surveys and workshops, the study used targeted expert interviews to identify research priorities in applying AI to AMR. Despite this abridged process, the extensive subsequent thematic analysis maintained the principles set out by the National Institute for Health and Care Research, which coordinates the infrastructure of the JLA to oversee the processes for PSPs, based at the NIHR Coordinating Centre, University of Southampton.
The BARDI framework advances previous antimicrobial-resistance proposals by transforming disconnected objectives into a unified, action-oriented ecosystem. Whereas previous action plans simply urged stronger surveillance, more research, better diagnostics, expanded R&D and the development of an economic case for investment, BARDI weaves these goals into five interlocking pillars that actively drive progress. First, instead of leaving data systems in siloed national or sectoral hands, BARDI establishes a neutral data broker. This broker enforces common standards, mediates access and protects proprietary interests, ensuring that human health, veterinary, agricultural and environmental datasets flow together under fair governance. Second, BARDI elevates AI from an aspirational research tool to the central engine of the response: AI-driven modelling continuously ingests brokered data to forecast resistance trends, optimise pharmacokinetic/pharmacodynamic regimens and even propose novel therapeutic compounds. Third, rapid diagnostics are promoted from a side objective to a full pillar. Decentralised, POC tests feed real-time results into the data broker, triggering AI-informed stewardship interventions and improving surveillance with minimal delay. Fourth, the drug-discovery pillar couples AI-identified bacterial targets with a prioritisation schema linked directly to public health impact, ensuring that investment goes toward the most critical vaccines and antimicrobial compounds rather than dispersing funding across undifferentiated R&D. Finally, integrated economic prevention converts the longstanding call to ‘develop the economic case’ into a dynamic funding mechanism. Real-time data on resistance burden and projected economic losses guide the allocation of resources to stewardship programmes, awareness campaigns, incentive prizes and cross-sector collaborations. In this way, economics is not merely a justification for action but an operational tool that continually sustains and adapts the entire BARDI ecosystem.
The top ten expert research themes for the application of AI to AMR provide valuable clarity on allocating resources to inform research developments in biosecurity and pandemic preparation. We propose a BARDI framework for directing AMR policy (Brokered data-sharing, AI-driven modelling, Rapid diagnostics, Drug discovery and Integrated economic prevention). Data was the most consistently raised issue throughout the interviews and needs to be neutrally coordinated to avoid obstacles, including automation bias, fear of job displacement and scepticism about AI’s decision-making abilities. Developing AI for AMR faces complex challenges, particularly in ensuring that AI tools comply with medical device regulations and data protection laws across different countries. The BARDI framework is a coherent strategy to fight AMR with AI to help realise the aspiration of the GRAM Project to save up to 92 million lives by 20503.
-

 Business3 days ago
Business3 days agoThe Guardian view on Trump and the Fed: independence is no substitute for accountability | Editorial
-
Tools & Platforms3 weeks ago
Building Trust in Military AI Starts with Opening the Black Box – War on the Rocks
-

 Ethics & Policy1 month ago
Ethics & Policy1 month agoSDAIA Supports Saudi Arabia’s Leadership in Shaping Global AI Ethics, Policy, and Research – وكالة الأنباء السعودية
-

 Events & Conferences3 months ago
Events & Conferences3 months agoJourney to 1000 models: Scaling Instagram’s recommendation system
-

 Jobs & Careers2 months ago
Jobs & Careers2 months agoMumbai-based Perplexity Alternative Has 60k+ Users Without Funding
-

 Funding & Business2 months ago
Funding & Business2 months agoKayak and Expedia race to build AI travel agents that turn social posts into itineraries
-

 Education2 months ago
Education2 months agoVEX Robotics launches AI-powered classroom robotics system
-

 Podcasts & Talks2 months ago
Podcasts & Talks2 months agoHappy 4th of July! 🎆 Made with Veo 3 in Gemini
-

 Podcasts & Talks2 months ago
Podcasts & Talks2 months agoOpenAI 🤝 @teamganassi
-

 Mergers & Acquisitions2 months ago
Mergers & Acquisitions2 months agoDonald Trump suggests US government review subsidies to Elon Musk’s companies