AI Research
AI text-to-speech programs could “unlearn” how to imitate certain people

AI companies generally keep a tight grip on their models to discourage misuse. For example, if you ask ChatGPT to give you someone’s phone number or instructions for doing something illegal, it will likely just tell you it cannot help. However, as many examples over time have shown, clever prompt engineering or model fine-tuning can sometimes get these models to say things they otherwise wouldn’t. The unwanted information may still be hiding somewhere inside the model so that it can be accessed with the right techniques.
At present, companies tend to deal with this issue by applying guardrails; the idea is to check whether the prompts or the AI’s responses contain disallowed material. Machine unlearning instead asks whether an AI can be made to forget a piece of information that the company doesn’t want it to know. The technique takes a leaky model and the specific training data to be redacted and uses them to create a new model—essentially, a version of the original that never learned that piece of data. While machine unlearning has ties to older techniques in AI research, it’s only in the past couple of years that it’s been applied to large language models.
Jinju Kim, a master’s student at Sungkyunkwan University who worked on the paper with Ko and others, sees guardrails as fences around the bad data put in place to keep people away from it. “You can’t get through the fence, but some people will still try to go under the fence or over the fence,” says Kim. But unlearning, she says, attempts to remove the bad data altogether, so there is nothing behind the fence at all.
The way current text-to-speech systems are designed complicates this a little more, though. These so-called “zero-shot” models use examples of people’s speech to learn to re-create any voice, including those not in the training set—with enough data, it can be a good mimic when supplied with even a small sample of someone’s voice. So “unlearning” means a model not only needs to “forget” voices it was trained on but also has to learn not to mimic specific voices it wasn’t trained on. All the while, it still needs to perform well for other voices.
To demonstrate how to get those results, Kim taught a recreation of VoiceBox, a speech generation model from Meta, that when it was prompted to produce a text sample in one of the voices to be redacted, it should instead respond with a random voice. To make these voices realistic, the model “teaches” itself using random voices of its own creation.
According to the team’s results, which are to be presented this week at the International Conference on Machine Learning, prompting the model to imitate a voice it has “unlearned” gives back a result that—according to state-of-the-art tools that measure voice similarity—mimics the forgotten voice more than 75% less effectively than the model did before. In practice, this makes the new voice unmistakably different. But the forgetfulness comes at a cost: The model is about 2.8% worse at mimicking permitted voices. While these percentages are a bit hard to interpret, the demo the researchers released online offers very convincing results, both for how well redacted speakers are forgotten and how well the rest are remembered. A sample from the demo is given below.
Ko says the unlearning process can take “several days,” depending on how many speakers the researchers want the model to forget. Their method also requires an audio clip about five minutes long for each speaker whose voice is to be forgotten.
In machine unlearning, pieces of data are often replaced with randomness so that they can’t be reverse-engineered back to the original. In this paper, the randomness for the forgotten speakers is very high—a sign, the authors claim, that they are truly forgotten by the model.
“I have seen people optimizing for randomness in other contexts,” says Vaidehi Patil, a PhD student at the University of North Carolina at Chapel Hill who researches machine unlearning. “This is one of the first works I’ve seen for speech.” Patil is organizing a machine unlearning workshop affiliated with the conference, and the voice unlearning research will also be presented there.
AI Research
MAIA platform for routine clinical testing: an artificial intelligence embryo selection tool developed to assist embryologists
Graham, M. E. et al. Assisted reproductive technology: Short- and long-term outcomes. Dev. Med. Child. Neurol. 65, 38–49 (2023).
Jiang, V. S. & Bormann, C. L. Artificial intelligence in the in vitro fertilization laboratory: a review of advancements over the last decade. Fertil. Steril. 120, 17–23 (2023).
Devine, K. et al. Single vitrified blastocyst transfer maximizes liveborn children per embryo while minimizing preterm birth. Fertil. Steril. 103, 1454–1460 (2015).
Tiitinen, A. Single embryo transfer: why and how to identify the embryo with the best developmental potential. Best Pract. Res. Clin. Endocrinol. Metab. 33, 77–88 (2019).
Glatstein, I., Chavez-Badiola, A. & Curchoe, C. L. New frontiers in embryo selection. J. Assist. Reprod. Genet. 40, 223–234 (2023).
Gardner, D. K. & Schoolcraft, W. B. Culture and transfer of human blastocysts. Curr. Opin. Obstet. Gynaecol. 11, 307–311 (1999).
Sciorio, R. & Meseguer, M. Focus on time-lapse analysis: blastocyst collapse and morphometric assessment as new features of embryo viability. Reprod. BioMed. Online. 43, 821–832 (2021).
Sundvall, L., Ingerslev, H. J., Knudsen, U. B. & Kirkegaard, K. Inter- and intra-observer variability of time-lapse annotations. Hum. Reprod. 28, 3215–3221 (2013).
Gallego, R. D., Remohí, J. & Meseguer, M. Time-lapse imaging: the state of the Art. Biol. Reprod. 101, 1146–1154 (2019).
VerMilyea, M. D. et al. Computer-automated time-lapse analysis results correlate with embryo implantation and clinical pregnancy: a blinded, multi-centre study. Reprod. Biomed. Online. 29, 729–736 (2014).
Chéles, D. S., Molin, E. A. D., Rocha, J. C. & Nogueira, M. F. G. Mining of variables from embryo morphokinetics, blastocyst’s morphology and patient parameters: an approach to predict the live birth in the assisted reproduction service. JBRA Assist. Reprod. 24, 470–479 (2020).
Rocha, C., Nogueira, M. G., Zaninovic, N. & Hickman, C. Is AI assessment of morphokinetic data and digital image analysis from time-lapse culture predictive of implantation potential of human embryos? Fertil. Steril. 110, e373 (2018).
Zaninovic, N. et al. Application of artificial intelligence technology to increase the efficacy of embryo selection and prediction of live birth using human blastocysts cultured in a time-lapse incubator. Fertil. Steril. 110, e372–e373 (2018).
Alegre, L. et al. First application of artificial neuronal networks for human live birth prediction on Geri time-lapse monitoring system blastocyst images. Fertil. Steril. 114, e140 (2020).
Bori, L. et al. An artificial intelligence model based on the proteomic profile of euploid embryos and blastocyst morphology: a preliminary study. Reprod. BioMed. Online. 42, 340–350 (2021).
Chéles, D. S. et al. An image processing protocol to extract variables predictive of human embryo fitness for assisted reproduction. Appl. Sci. 12, 3531 (2022).
Jacobs, C. K. et al. Embryologists versus artificial intelligence: predicting clinical pregnancy out of a transferred embryo who performs it better? Fertil. Steril. 118, e81–e82 (2022).
Lorenzon, A. et al. P-211 development of an artificial intelligence software with consistent laboratory data from a single IVF center: performance of a new interface to predict clinical pregnancy. Hum. Reprod. 39, deae108.581 (2024).
Fernandez, E. I. et al. Artificial intelligence in the IVF laboratory: overview through the application of different types of algorithms for the classification of reproductive data. J. Assist. Reprod. Genet. 37, 2359–2376 (2020).
Mendizabal-Ruiz, G. et al. Computer software (SiD) assisted real-time single sperm selection associated with fertilization and blastocyst formation. Reprod. BioMed. Online. 45, 703–711 (2022).
Fjeldstad, J. et al. Segmentation of mature human oocytes provides interpretable and improved blastocyst outcome predictions by a machine learning model. Sci. Rep. 14, 10569 (2024).
Khosravi, P. et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit. Med. 2, 21 (2019).
Hickman, C. et al. Inner cell mass surface area automatically detected using Chloe eq™(fairtility), an ai-based embryology support tool, is associated with embryo grading, embryo ranking, ploidy and live birth outcome. Fertil. Steril. 118, e79 (2022).
Tran, D., Cooke, S., Illingworth, P. J. & Gardner, D. K. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum. Reprod. 34, 1011–1018 (2019).
Rajendran, S. et al. Automatic ploidy prediction and quality assessment of human blastocysts using time-lapse imaging. Nat. Commun. 15, 7756 (2024).
Bormann, C. L. et al. Consistency and objectivity of automated embryo assessments using deep neural networks. Fertil. Steril. 113, 781–787e1 (2020).
Kragh, M. F. & Karstoft, H. Embryo selection with artificial intelligence: how to evaluate and compare methods? J. Assist. Reprod. Genet. 38, 1675–1689 (2021).
Cromack, S. C., Lew, A. M., Bazzetta, S. E., Xu, S. & Walter, J. R. The perception of artificial intelligence and infertility care among patients undergoing fertility treatment. J. Assist. Reprod. Genet. https://doi.org/10.1007/s10815-024-03382-5 (2025).
Fröhlich, H. et al. From hype to reality: data science enabling personalized medicine. BMC Med. 16, 150 (2018).
Zhu, J. et al. External validation of a model for selecting day 3 embryos for transfer based upon deep learning and time-lapse imaging. Reprod. BioMed. Online. 47, 103242 (2023).
Yelke, H. K. et al. O-007 Simplifying the complexity of time-lapse decisions with AI: CHLOE (Fairtility) can automatically annotate morphokinetics and predict blastulation (at 30hpi), pregnancy and ongoing clinical pregnancy. Hum. Reprod. 37, deac104.007 (2022).
Papatheodorou, A. et al. Clinical and practical validation of an end-to-end artificial intelligence (AI)-driven fertility management platform in a real-world clinical setting. Reprod. BioMed. Online. 45, e44–e45 (2022).
Salih, M. et al. Embryo selection through artificial intelligence versus embryologists: a systematic review. Hum. Reprod. Open hoad031 (2023).
Nunes, K. et al. Admixture’s impact on Brazilian population evolution and health. Science. 388(6748), eadl3564 (2025).
Jackson-Bey, T. et al. Systematic review of Racial and ethnic disparities in reproductive endocrinology and infertility: where do we stand today? F&S Reviews. 2, 169–188 (2021).
Kassi, L. A. et al. Body mass index, not race, May be associated with an alteration in early embryo morphokinetics during in vitro fertilization. J. Assist. Reprod. Genet. 38, 3091–3098 (2021).
Pena, S. D. J., Bastos-Rodrigues, L., Pimenta, J. R. & Bydlowski, S. P. DNA tests probe the genomic ancestry of Brazilians. Braz J. Med. Biol. Res. 42, 870–876 (2009).
Fraga, A. M. et al. Establishment of a Brazilian line of human embryonic stem cells in defined medium: implications for cell therapy in an ethnically diverse population. Cell. Transpl. 20, 431–440 (2011).
Amin, F. & Mahmoud, M. Confusion matrix in binary classification problems: a step-by-step tutorial. J. Eng. Res. 6, 0–0 (2022).
Magdi, Y. et al. Effect of embryo selection based morphokinetics on IVF/ICSI outcomes: evidence from a systematic review and meta-analysis of randomized controlled trials. Arch. Gynecol. Obstet. 300, 1479–1490 (2019).
Guo, Y. H., Liu, Y., Qi, L., Song, W. Y. & Jin, H. X. Can time-lapse incubation and monitoring be beneficial to assisted reproduction technology outcomes? A randomized controlled trial using day 3 double embryo transfer. Front. Physiol. 12, 794601 (2022).
Giménez, C., Conversa, L., Murria, L. & Meseguer, M. Time-lapse imaging: morphokinetic analysis of in vitro fertilization outcomes. Fertil. Steril. 120, 228–227 (2023).
Vitrolife EmbryoScope + time-lapse system. (2023). https://www.vitrolife.com/products/time-lapse-systems/embryoscopeplus-time-lapse-system/.
Lagalla, C. et al. A quantitative approach to blastocyst quality evaluation: morphometric analysis and related IVF outcomes. J. Assist. Reprod. Genet. 32, 705–712 (2015).
Rocha, J. C. et al. A method based on artificial intelligence to fully automatize the evaluation of bovine blastocyst images. Sci. Rep. 7, 7659 (2017).
Chavez-Badiola, A. et al. Predicting pregnancy test results after embryo transfer by image feature extraction and analysis using machine learning. Sci. Rep. 10, 4394 (2020).
Matos, F. D., Rocha, J. C. & Nogueira, M. F. G. A method using artificial neural networks to morphologically assess mouse blastocyst quality. J. Anim. Sci. Technol. 56, 15 (2014).
Wang, S., Zhou, C., Zhang, D., Chen, L. & Sun, H. A deep learning framework design for automatic blastocyst evaluation with multifocal images. IEEE Access. 9, 18927–18934 (2021).
Berntsen, J., Rimestad, J., Lassen, J. T., Tran, D. & Kragh, M. F. Robust and generalizable embryo selection based on artificial intelligence and time-lapse image sequences. PLoS One. 17, e0262661 (2022).
Fruchter-Goldmeier, Y. et al. An artificial intelligence algorithm for automated blastocyst morphometric parameters demonstrates a positive association with implantation potential. Sci. Rep. 13, 14617 (2023).
Illingworth, P. J. et al. Deep learning versus manual morphology-based embryo selection in IVF: a randomized, double-blind noninferiority trial. Nat. Med. 30, 3114–3120 (2024).
Kanakasabapathy, M. K. et al. Development and evaluation of inexpensive automated deep learning-based imaging systems for embryology. Lab. Chip. 19, 4139–4145 (2019).
Loewke, K. et al. Characterization of an artificial intelligence model for ranking static images of blastocyst stage embryos. Fertil. Steril. 117, 528–535 (2022).
Hengstschläger, M. Artificial intelligence as a door opener for a new era of human reproduction. Hum. Reprod. Open hoad043 (2023).
Lassen Theilgaard, J., Fly Kragh, M., Rimestad, J., Nygård Johansen, M. & Berntsen, J. Development and validation of deep learning based embryo selection across multiple days of transfer. Sci. Rep. 13 (1), 4235 (2023).
Lozano, M. et al. P-301 Assessment of ongoing clinical outcomes prediction of an AI system on retrospective SET data, Human Reprod. 38(Issue Supplement_1), dead093.659. (2023).
Collins, G. S. et al. TRIPOD + AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ 385, e078378 (2024).
Abdolrasol, M. G. M. et al. Artificial neural networks based optimization techniques: a review. Electronics 10, 2689 (2021).
Yuzer, E. O. & Bozkurt, A. Instant solar irradiation forecasting for solar power plants using different ANN algorithms and network models. Electr. Eng. 106, 3671–3689 (2024).
Guariso, G. & Sangiorgio, M. Improving the performance of multiobjective genetic algorithms: an elitism-based approach. Information 11, 587 (2020).
García-Pascual, C. M. et al. Optimized NGS approach for detection of aneuploidies and mosaicism in PGT-A and imbalances in PGT-SR. Genes 11, 724 (2020).
AI Research
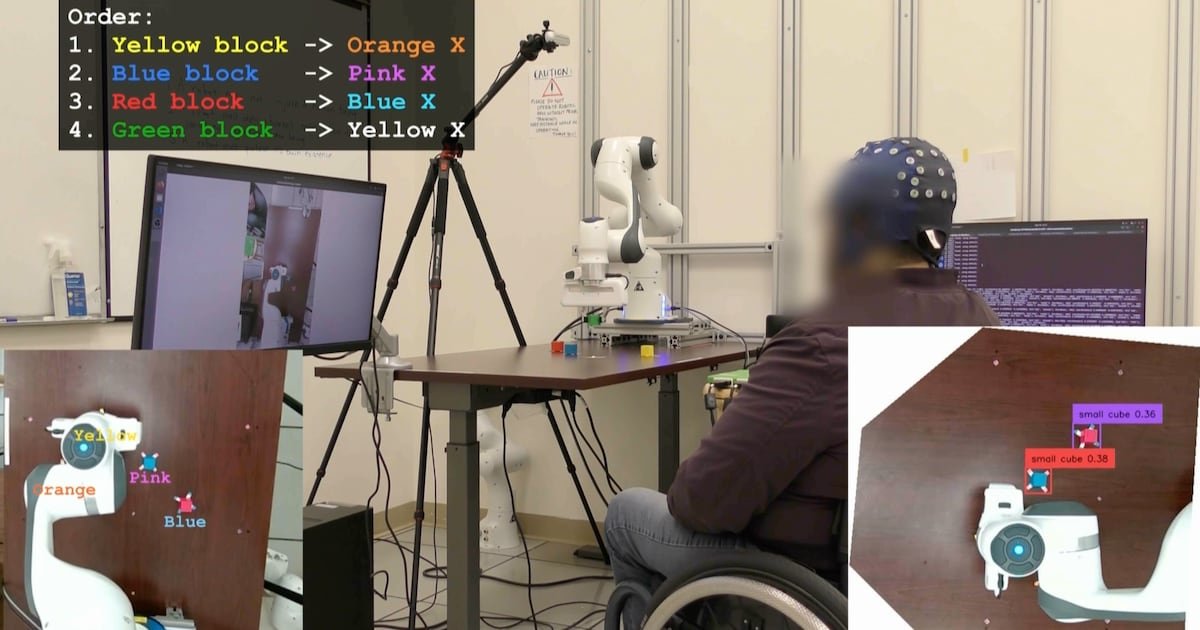
UCLA Researchers Enable Paralyzed Patients to Control Robots with Thoughts Using AI – CHOSUNBIZ – Chosun Biz
AI Research
Hackers exploit hidden prompts in AI images, researchers warn

Cybersecurity firm Trail of Bits has revealed a technique that embeds malicious prompts into images processed by large language models (LLMs). The method exploits how AI platforms compress and downscale images for efficiency. While the original files appear harmless, the resizing process introduces visual artifacts that expose concealed instructions, which the model interprets as legitimate user input.
In tests, the researchers demonstrated that such manipulated images could direct AI systems to perform unauthorized actions. One example showed Google Calendar data being siphoned to an external email address without the user’s knowledge. Platforms affected in the trials included Google’s Gemini CLI, Vertex AI Studio, Google Assistant on Android, and Gemini’s web interface.
Read More: Meta curbs AI flirty chats, self-harm talk with teens
The approach builds on earlier academic work from TU Braunschweig in Germany, which identified image scaling as a potential attack surface in machine learning. Trail of Bits expanded on this research, creating “Anamorpher,” an open-source tool that generates malicious images using interpolation techniques such as nearest neighbor, bilinear, and bicubic resampling.
From the user’s perspective, nothing unusual occurs when such an image is uploaded. Yet behind the scenes, the AI system executes hidden commands alongside normal prompts, raising serious concerns about data security and identity theft. Because multimodal models often integrate with calendars, messaging, and workflow tools, the risks extend into sensitive personal and professional domains.
Also Read: Nvidia CEO Jensen Huang says AI boom far from over
Traditional defenses such as firewalls cannot easily detect this type of manipulation. The researchers recommend a combination of layered security, previewing downscaled images, restricting input dimensions, and requiring explicit confirmation for sensitive operations.
“The strongest defense is to implement secure design patterns and systematic safeguards that limit prompt injection, including multimodal attacks,” the Trail of Bits team concluded.
-

 Business3 days ago
Business3 days agoThe Guardian view on Trump and the Fed: independence is no substitute for accountability | Editorial
-
Tools & Platforms3 weeks ago
Building Trust in Military AI Starts with Opening the Black Box – War on the Rocks
-

 Ethics & Policy1 month ago
Ethics & Policy1 month agoSDAIA Supports Saudi Arabia’s Leadership in Shaping Global AI Ethics, Policy, and Research – وكالة الأنباء السعودية
-

 Events & Conferences3 months ago
Events & Conferences3 months agoJourney to 1000 models: Scaling Instagram’s recommendation system
-

 Jobs & Careers2 months ago
Jobs & Careers2 months agoMumbai-based Perplexity Alternative Has 60k+ Users Without Funding
-

 Funding & Business2 months ago
Funding & Business2 months agoKayak and Expedia race to build AI travel agents that turn social posts into itineraries
-

 Education2 months ago
Education2 months agoVEX Robotics launches AI-powered classroom robotics system
-

 Podcasts & Talks2 months ago
Podcasts & Talks2 months agoHappy 4th of July! 🎆 Made with Veo 3 in Gemini
-

 Podcasts & Talks2 months ago
Podcasts & Talks2 months agoOpenAI 🤝 @teamganassi
-

 Mergers & Acquisitions2 months ago
Mergers & Acquisitions2 months agoDonald Trump suggests US government review subsidies to Elon Musk’s companies