AI Research
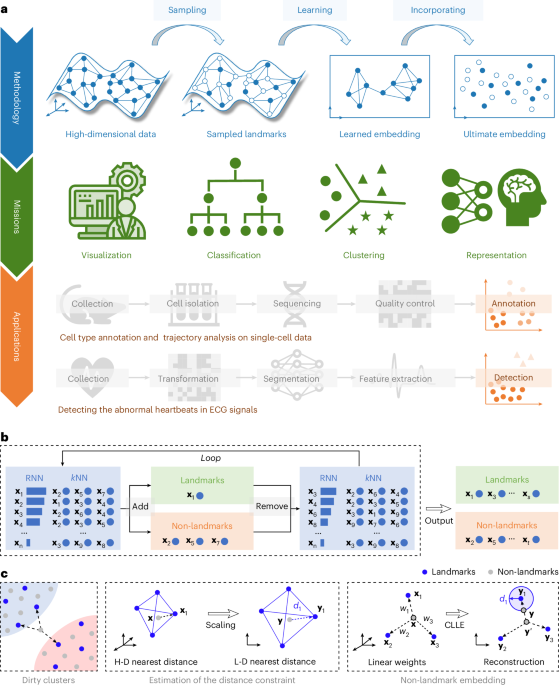
Sampling-enabled scalable manifold learning unveils the discriminative cluster structure of high-dimensional data
Busch, E. L. et al. Multi-view manifold learning of human brain-state trajectories. Nat. Comput. Sci. 3, 240–253 (2023).
Floryan, D. & Graham, M. D. Data-driven discovery of intrinsic dynamics. Nat. Mach. Intell. 4, 1113–1120 (2022).
Islam, M. T. et al. Revealing hidden patterns in deep neural network feature space continuum via manifold learning. Nat. Commun. 14, 8506 (2023).
Vogelstein, J. T. et al. Supervised dimensionality reduction for big data. Nat. Commun. 12, 2872 (2021).
Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 25, 417–441 (1933).
Peng, D., Gui, Z., Gui, J., & Wu, H. A robust and efficient boundary point detection method by measuring local direction dispersion. IEEE Trans. Circuits Syst. Video Technol. https://doi.org/10.1109/TCSVT.2025.3546401 (2025).
Kruskal, J. B. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 29, 1–27 (1964).
Fisher, R. A. The use of multiple measurements in taxonomic problems. Ann. Eugenics 7, 179–188 (1936).
Yan, S. et al. Nonlinear discriminant analysis on embedded manifold. IEEE Trans. Circuits Syst. Video Technol. 17, 468–477 (2007).
Zhang, L., Zhou, W. & Chang, P.-C. Generalized nonlinear discriminant analysis and its small sample size problems. Neurocomputing 74, 568–574 (2011).
Tenenbaum, J. B., Silva, V. D. & Langford, J. C. A global geometric framework for nonlinear dimensionality reduction. Science 290, 2319–2323 (2000).
Roweis, S. T. & Saul, L. K. Nonlinear dimensionality reduction by locally linear embedding. Science 290, 2323–2326 (2000).
Cover, T. & Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 13, 21–27 (1967).
Belkin, M., & Niyogi, P. Laplacian eigenmaps and spectral techniques for embedding and clustering. In Proc. Advanced Neural Information Processing Systems (eds Dietterich, T. G. et al.) 585–591 (2001).
Donoho, D. L. & Grimes, C. Hessian eigenmaps: Locally linear embedding techniques for high-dimensional data. Proc. Natl Acad. Sci. USA 100, 5591–5596 (2003).
Coifman, R. R. et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc. Natl Acad. Sci. USA 102, 7426–7431 (2005).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
McInnes, L., Healy, J., Saul, N. & Groçberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3, 861 (2018).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Amid, E. & Warmuth, M. K. TriMap: large-scale dimensionality reduction using triplets. Preprint at https://arxiv.org/abs/1910.00204 (2022).
Fu, C., Zhang, Y., Cai, D. & Ren, X. AtSNE: efficient and robust visualization on GPU through hierarchical optimization. In Proc. 25th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD) 176–186 (Association for Computing Machinery, 2019).
Moon, K. R. et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482–1492 (2019).
Kobak, D. & Berens, P. The art of using t-SNE for single-cell transcriptomics. Nat. Commun. 10, 5416 (2019).
Shen, C. & Wu, H.-T. Scalability and robustness of spectral embedding: landmark diffusion is all you need. Inf. Inference 11, 1527–1595 (2022).
Long, A. W. & Ferguson, A. L. Landmark diffusion maps (L-dMaps): accelerated manifold learning out-of-sample extension. Appl. Comput. Harmon. Anal. 47, 190–211 (2019).
Persad, S. et al. SEACells infers transcriptional and epigenomic cellular states from single-cell genomics data. Nat. Biotechnol. 41, 1746–1757 (2023).
Stanoi, I., Riedewald, M., Agrawal, D. & El Abbadi, A. Discovery of influence sets in frequently updated databases. In Proc. 27th International Conference on Very Large Databases (VLDB) (eds. Apers P. M. G. et al.) 99–108 (Morgan Kaufmann Publishers Inc., 2001).
Tao, Y., Papadias, D. & Lian, X. Reverse kNN search in arbitrary dimensionality. In Proc. 30th International Conference on Very Large Databases (VLDB) (eds Nascimento, M. A. et al.) 744–755 (Morgan Kaufmann Publishers Inc., 2004).
Radovanovic, M., Nanopoulos, A. & Ivanovic, M. Hubs in space: popular nearest neighbors in high-dimensional data. J. Mach. Learn. Res. 11, 2487–2531 (2010).
Loshchilov, I. & Hutter, F. SGDR: stochastic gradient descent with warm restarts. In Proc. 5th International Conference on Learning Representations (ICLR) 1769–1784 (Curran Associates, 2017).
Peng, D., Gui, Z. & Wu, H. MeanCut: a greedy-optimized graph clustering via path-based similarity and degree descent criterion. Preprint at https://arxiv.org/abs/2312.04067 (2023).
Karypis, G., Han, E.-H. & Kumar, V. Chameleon: hierarchical clustering using dynamic modeling. Computer 32, 68–75 (1999).
France, S. & Carroll, D. in Machine Learning and Data Mining in Pattern Recognition (ed. Perner, P.) vol. 4,571, 499–517 (2007).
Moor, M., Horn, M., Rieck, B. & Borgwardt, K. Topological autoencoders. In Proc. 37th International Conference on Machine Learning (ICML) (eds Daumé, H. & Singh, A.) 7045–7054 (PMLR, 2020).
Sainburg, T., McInnes, L. & Gentner, T. Q. Parametric UMAP embeddings for representation and semisupervised learning. Neural Comput. 33, 2881–2907 (2021).
Dua, D. & Graff, C. UCI machine learning repository. University of California Irvine https://archive.ics.uci.edu (2019).
Krizhevsky, A. & Hinton, G. Learning Multiple Layers of Features from Tiny Images Technical Report (Univ. Toronto, 2009).
Lecun, Y., Bottou, L., Bengio, Y. & Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998).
Xiao, H., Rasul, K. & Vollgraf, R. Fashion-MNIST: a novel image dataset for benchmarking machine learning algorithms. Preprint at https://arxiv.org/abs/1708.07747 (2017).
Corso, G. M. D., Gulli, A. & Romani, F. Ranking a stream of news. In Proc. 14th International World Wide Web Conference (WWW) 97–106 (Association for Computing Machinery, 2005).
Poličar, P. G., Stražar, M. & Zupan, B. openTSNE: a modular Python library for t-SNE dimensionality reduction and embedding. J. Stat. Softw. 109, 1–30 (2024).
Huang, H., Wang, Y., Rudin, C. & Browne, E. P. Towards a comprehensive evaluation of dimension reduction methods for transcriptomic data visualization. Commun. Biol. 5, 719 (2022).
Amir, E. A. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
Khan, S. A. et al. Reusability report: learning the transcriptional grammar in single-cell RNA-sequencing data using transformers. Nat. Mach. Intell. 5, 1437–1446 (2023).
Heng, J. S. et al. Hypoxia tolerance in the Norrin-deficient retina and the chronically hypoxic brain studied at single-cell resolution. Proc. Natl Acad. Sci. USA 116, 9103–9114 (2019).
Satija, R. et al. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Kiselev, V. Y., Andrews, T. S. & Hemberg, M. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 20, 273–282 (2019).
Peng, D. et al. Clustering by measuring local direction centrality for data with heterogeneous density and weak connectivity. Nat. Commun. 13, 5455 (2022).
Wang, S. K. et al. Single-cell multiome and enhancer connectome of human retinal pigment epithelium and choroid nominate pathogenic variants in age-related macular degeneration. Preprint at bioRxiv https://doi.org/10.1101/2025.03.21.644670 (2025).
Ziegler, C. G. et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell 184, 4713–4733 (2021).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Weber, L. M. & Robinson, M. D. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytom. A 89, 1084–1096 (2016).
Ferrari, G., Thrasher, A. J. & Aiuti, A. Gene therapy using haematopoietic stem and progenitor cells. Nat. Rev. Genet. 22, 216–234 (2021).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Young, W. J. et al. Genetic analyses of the electrocardiographic QT interval and its components identify additional loci and pathways. Nat. Commun. 13, 5144 (2022).
Niroshana, S. M. I., Kuroda, S., Tanaka, K. & Chen, W. Beat-wise segmentation of electrocardiogram using adaptive windowing and deep neural network. Sci. Rep. 13, 11039 (2023).
Huo, R. et al. ECG segmentation algorithm based on bidirectional hidden semi-Markov model. Comput. Biol. Med. 150, 106081 (2022).
Oberlin, T., Meignen, S. & Perrier, V. The Fourier-based synchrosqueezing transform. In Proc. IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 315–319 (IEEE, 2014).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
Laguna, P., Mark, R. G., Goldberg, A. & Moody, G. B. A database for evaluation of algorithms for measurement of QT and other waveform intervals in the ECG. Comput. Cardiol. 24, 673–676 (1997).
Goldberger, A. L. et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101, e215–e220 (2000).
Bengio, Y., Courville, A. & Vincent, P. Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1798–1828 (2013).
Peng, D., Gui, Z. & Wu, H. Interpreting the curse of dimensionality from distance concentration and manifold effect. Preprint at https://arxiv.org/abs/2401.00422 (2023).
Healy, J. & McInnes, L. Uniform manifold approximation and projection. Nat. Rev. Methods Primers 4, 82 (2024).
Malkov, Y. A. & Yashunin, D. A. Efficient and robust approximate nearest neighbor search using hierarchical navigable small world graphs. IEEE Trans. Pattern Anal. Mach. Intell. 42, 824–836 (2018).
Dong, W., Moses, C. & Li, K. Efficient k-nearest neighbor graph construction for generic similarity measures. In Proc. 20th International Conference on World Wide Web (WWW) 577–586 (Association for Computing Machinery, 2011).
Saul, L. K. & Roweis, S. T. Think globally, fit locally: unsupervised learning of low dimensional manifolds. J. Mach. Learn. Res. 4, 119–155 (2003).
Wang, J. Real local-linearity preserving embedding. Neurocomputing 136, 7–13 (2014).
Wang, Z. et al. Clustering by local gravitation. IEEE Trans. Cybern. 48, 1383–1396 (2018).
von Luxburg, U. A tutorial on spectral clustering. Stat. Comp. 17, 395–416 (2007).
Ding, J., Shah, S. & Condon, A. densityCut: an efficient and versatile topological approach for automatic clustering of biological data. Bioinformatics 32, 2567–2576 (2016).
Kuhn, H. W. The Hungarian method for the assignment problem. Nav. Res. Logist. Q. 2, 83–97 (1955).
Peng, D. Source code of SUDE. Zenodo https://doi.org/10.5281/zenodo.16792257 (2025).
Peng, D. Open code for in-review “Sampling-enabled scalable manifold learning unveils discriminative cluster structure of high-dimensional data”. Code Ocean https://doi.org/10.24433/CO.9866909.v3 (2025).
AI Research
Will artificial intelligence fuel moral chaos or positive change?

Artificial intelligence is transforming our world at an unprecedented rate, but what does this mean for Christians, morality and human flourishing?
In this episode of “The Inside Story,” Billy Hallowell sits down with The Christian Post’s Brandon Showalter to unpack the promises and perils of AI.
From positives like Bible translation to fears over what’s to come, they explore how believers can apply a biblical worldview to emerging technology, the dangers of becoming “subjects” of machines, and why keeping Christ at the center is the only true safeguard.
Plus, learn about The Christian Post’s upcoming “AI for Humanity” event at Colorado Christian University and how you can join the conversation in person or via livestream:
“The Inside Story” takes you behind the headlines of the biggest faith, culture and political headlines of the week. In 15 minutes or less, Christian Post staff writers and editors will help you navigate and understand what’s driving each story, the issues at play — and why it all matters.
Listen to more Christian podcasts today on the Edifi app — and be sure to subscribe to The Inside Story on your favorite platforms:
AI Research
BNY and Carnegie Mellon University announce five-year $10 million partnership supporting AI research — EdTech Innovation Hub

The $10 million deal aims to bring students, faculty and staff together alongside BNY experts to advance AI applications and systems to prepare the next generation of leaders.
Known as the BNY AI Lab, the collaboration will focus on technologies and frameworks that can ensure robust governance of mission-critical AI applications.
“As AI drives productivity, unlocks growth and transforms industries, Pittsburgh has cemented its role as a global hub for innovation and talent, reinforcing Pennsylvania’s leadership in shaping the broader AI ecosystem,” comments Robin Vince, CEO at BNY. “Building on BNY’s 150-year legacy in the Commonwealth, we are proud to expand our work with Carnegie Mellon University to help attract world-class talent and pioneer AI research with an impact far beyond the region.”
A dedicated space for the collaboration will be created at the University’s Pittsburgh campus during the 2025-26 academic year.
“AI has emerged as one of the single most important intellectual developments of our time, and it is rapidly expanding into every sector of our economy,” adds Farnam Jahanian, President of Carnegie Mellon. “Carnegie Mellon University is thrilled to collaborate with BNY – a global financial services powerhouse – to responsibly develop and scale emerging AI technologies and democratize their impact for the benefit of industry and society at large.”
The ETIH Innovation Awards 2026
The EdTech Innovation Hub Awards celebrate excellence in global education technology, with a particular focus on workforce development, AI integration, and innovative learning solutions across all stages of education.
Now open for entries, the ETIH Innovation Awards 2026 recognize the companies, platforms, and individuals driving transformation in the sector, from AI-driven assessment tools and personalized learning systems, to upskilling solutions and digital platforms that connect learners with real-world outcomes.
Submissions are open to organizations across the UK, the Americas, and internationally. Entries should highlight measurable impact, whether in K–12 classrooms, higher education institutions, or lifelong learning settings.
Winners will be announced on 14 January 2026 as part of an online showcase featuring expert commentary on emerging trends and standout innovation. All winners and finalists will also be featured in our first print magazine, to be distributed at BETT 2026.
AI Research
Beyond Refusal — Constructive Safety Alignment for Responsible Language Models

View a PDF of the paper titled Oyster-I: Beyond Refusal — Constructive Safety Alignment for Responsible Language Models, by Ranjie Duan and 26 other authors
Abstract:Large language models (LLMs) typically deploy safety mechanisms to prevent harmful content generation. Most current approaches focus narrowly on risks posed by malicious actors, often framing risks as adversarial events and relying on defensive refusals. However, in real-world settings, risks also come from non-malicious users seeking help while under psychological distress (e.g., self-harm intentions). In such cases, the model’s response can strongly influence the user’s next actions. Simple refusals may lead them to repeat, escalate, or move to unsafe platforms, creating worse outcomes. We introduce Constructive Safety Alignment (CSA), a human-centric paradigm that protects against malicious misuse while actively guiding vulnerable users toward safe and helpful results. Implemented in Oyster-I (Oy1), CSA combines game-theoretic anticipation of user reactions, fine-grained risk boundary discovery, and interpretable reasoning control, turning safety into a trust-building process. Oy1 achieves state-of-the-art safety among open models while retaining high general capabilities. On our Constructive Benchmark, it shows strong constructive engagement, close to GPT-5, and unmatched robustness on the Strata-Sword jailbreak dataset, nearing GPT-o1 levels. By shifting from refusal-first to guidance-first safety, CSA redefines the model-user relationship, aiming for systems that are not just safe, but meaningfully helpful. We release Oy1, code, and the benchmark to support responsible, user-centered AI.
Submission history
From: Ranjie Duan [view email]
[v1]
Tue, 2 Sep 2025 03:04:27 UTC (5,745 KB)
[v2]
Thu, 4 Sep 2025 11:54:06 UTC (5,745 KB)
[v3]
Mon, 8 Sep 2025 15:18:35 UTC (5,746 KB)
[v4]
Fri, 12 Sep 2025 04:23:22 UTC (5,747 KB)
-

 Business2 weeks ago
Business2 weeks agoThe Guardian view on Trump and the Fed: independence is no substitute for accountability | Editorial
-
Tools & Platforms1 month ago
Building Trust in Military AI Starts with Opening the Black Box – War on the Rocks
-

 Ethics & Policy2 months ago
Ethics & Policy2 months agoSDAIA Supports Saudi Arabia’s Leadership in Shaping Global AI Ethics, Policy, and Research – وكالة الأنباء السعودية
-

 Events & Conferences4 months ago
Events & Conferences4 months agoJourney to 1000 models: Scaling Instagram’s recommendation system
-

 Jobs & Careers3 months ago
Jobs & Careers3 months agoMumbai-based Perplexity Alternative Has 60k+ Users Without Funding
-

 Podcasts & Talks2 months ago
Podcasts & Talks2 months agoHappy 4th of July! 🎆 Made with Veo 3 in Gemini
-

 Education3 months ago
Education3 months agoVEX Robotics launches AI-powered classroom robotics system
-

 Education2 months ago
Education2 months agoMacron says UK and France have duty to tackle illegal migration ‘with humanity, solidarity and firmness’ – UK politics live | Politics
-

 Podcasts & Talks2 months ago
Podcasts & Talks2 months agoOpenAI 🤝 @teamganassi
-

 Funding & Business3 months ago
Funding & Business3 months agoKayak and Expedia race to build AI travel agents that turn social posts into itineraries